 |
Plant Transcriptional Regulatory Map
|
FunTFBS
The tool screens for functional TFBS and regulatory interaction by coupling base-varied binding affinities of transcription factors and consistently evolutionary constraints on transcription factor binding sites.
Introduction to FunTFBS
FunTFBS is a tool used for identifying transcriptional factor binding sites (TFBS) which have transcriptional regulatory functions. According to binding motifs, the sequence affinities of TFs to genome are different among base pairs within binding sites. The mutations in different base pairs have different effects on the binding of TFs and thus the selective constrains are different. So for functional binding sites, the base pairs with higher affinity are more conserved across species than non-functional ones. Hinted by this, FunTFBS was developed to identify functional TFBS based on the correlation between frequencies in binding motifs and conservation scores. Given the genomic position of candidate TFBS, TF binding motif, genome sequence and PhyloP scores, FunTFBS can filter for functional ones based on the correlation between frequencies in binding motif and conservation scores across base pairs.
- Effective: This data from this method showed higher supported ratio by confirmed functional data with transcriptional regulatory functions than other motif based methods.
- Convenient: This method need only binding motifs, conservation data and genome sequences as input to filter for functional TFBS, which is free of other experimental data.
- Universal: This method is based on evolutionary constrains and thus could be used for other species.
- Rapid: This method can handle 10,000 candidate TFBS in less than 2 minutes (one cpu with 2.13GHz)
The functional TFBS in 63 plant species have been identified in our database, which covered all main branches of green plants.
Work Flow of FunTFBS
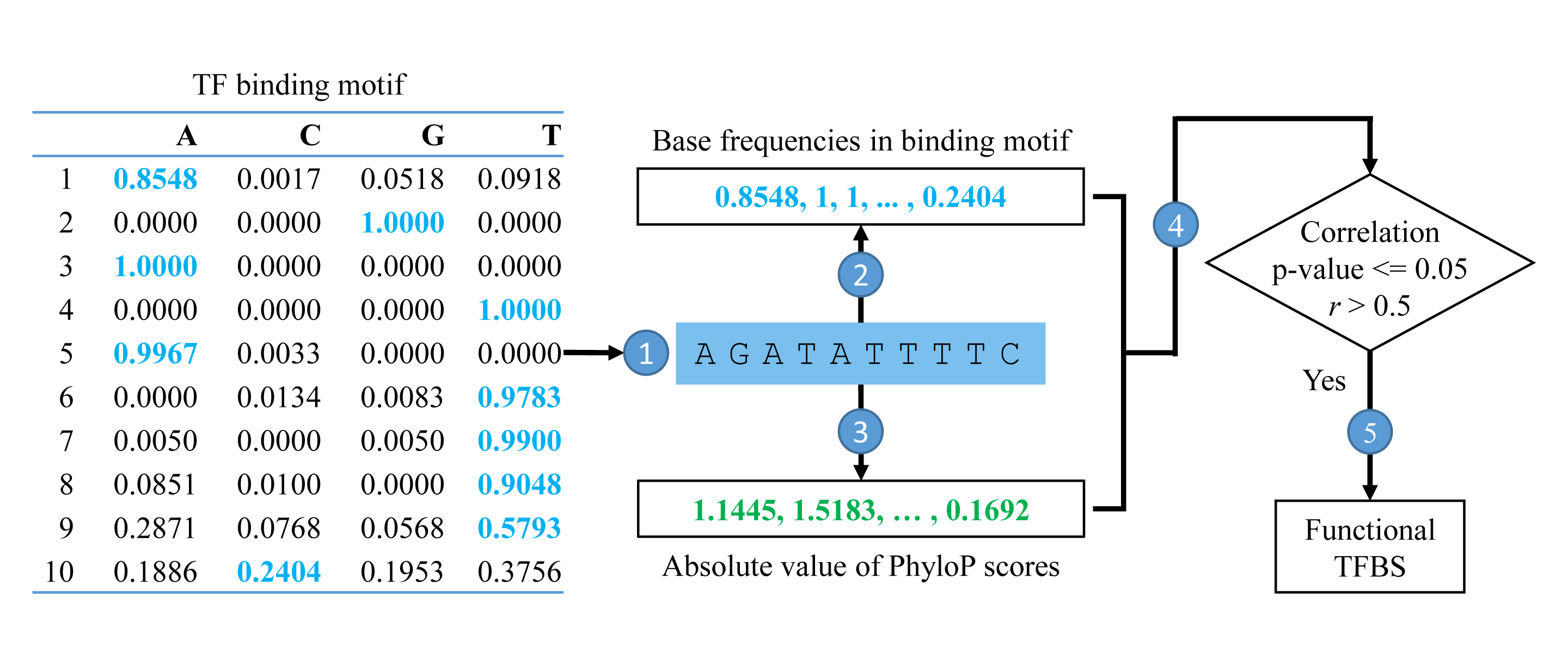
- Given the genomic position of specific TFBS, the genomic sequence is extracted.
- The motif frequencies of each base pair are extracted according to genomic sequence and the binding motif of corresponding TF.
- The PhyloP scores in the same region are extracted with the same order with motif frequencies (the PhyloP scores will be reversed for binding in negative strands).
- Pearson correlation is calculated between motif base frequencies and the absolute value of PhyloP scores.
- If the correlation test is significant (p<=0.05) with correlation score > 0.5, the corresponding binding site will be treated as functional TFBS.
Download
| Version | Date | Size | Link |
| 1.1.0 | 2018-03-25 | 255 kb | download |
Also you can download the pre-release from Github.
git clone https://github.com/gao-lab/FunTFBS.git
Installation
Prerequisite
- Perl (5.010 or later)
- R (3.0.1 or later)
- R package: data.table (1.10.4 or later)
Unpack the tarball
tar -zxvf funTFBS-1.1.0.tar.gz
Then go into the directory and the "funTFBS" file can be run directly.
cd funTFBS-1.1.0/ ./funTFBS
Also you can add this path to the PATH environment variable and run it out of directory:
export PATH=$PATH:/the path of this package/funTFBS-1.1.0 funTFBS
General usage
funTFBS -t TFBS.bed -m motifs -f motif-format -p PhyloP -g genome -o output
-t [TFBS.bed] the file containing positions of candidate TFBS in bed format.
-m [motifs] the file containing binding motifs in specified format.
-f [format] the format of bidning motifs.
-p [PhyloP.bg] the file containing PhyloP scores in bedGraph format.
-g [genome.fa] the file containing genomic sequence in fasta format.
-o [output] the output directory.
-h show this help information.
There are four parts of information for FunTFBS as input:
- Candidate TFBS in bed format with strand information, For example:
Chr1 3140 3161 AT3G27010 7.46e-06 + Chr1 3148 3162 AT5G08330 8.37e-06 - Chr1 3214 3229 AT2G20110 9.93e-06 + Chr1 3246 3258 AT1G69120 7.7e-06 + Chr1 3296 3311 AT2G32460 5.13e-06 +
Note: The 4th column of the file will be used as TF ID, which should be matched with TF ID in the motif file.
This file can be generated by ChIP-seq analysis or motif scanning. - Binding motif of TF
This file could be one of following formats:
meme/beeml/chen/jaspar-pfm/jaspar-sites/jaspar-cm/transfac/uniprobe
For example (meme format):
MEME version 4.4 ALPHABET= ACGT strands: + - Background letter frequencies (from file `../../promoter_background/Ath.bg'): A 0.33230 C 0.16770 G 0.16770 T 0.33230 MOTIF AT1G01060 MP00119 letter-probability matrix: alength= 4 w= 10 nsites= 599 E= 4.8e-803 0.854758 0.001669 0.051753 0.091820 0.000000 0.000000 1.000000 0.000000 1.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 1.000000 0.996661 0.003339 0.000000 0.000000 0.000000 0.013356 0.008347 0.978297 0.005008 0.000000 0.005008 0.989983 0.085142 0.010017 0.000000 0.904841 0.287145 0.076795 0.056761 0.579299 0.188648 0.240401 0.195326 0.375626 URL http://planttfdb.gao-lab.org/tf.php?sp=Ath&did=AT1G01060.1#bind_motif
Note: The TF ID should be matched with those in TFBS file (the 4th column), otherwise the motif base frequencies will not be extracted successfully.
This file can be generated by experiments (such as SELEX, PBM or ChIP-seq) or downloaded from TF motif database. - PhyloP score in bedGraph format, for example:
Chr1 116 117 0.263925266926064 Chr1 117 118 0.27368586219026 Chr1 118 119 -0.991320815703633 Chr1 119 120 0.27368586219026 Chr1 120 121 0.387503444300272 Chr1 121 122 0.27368586219026 Chr1 122 123 0.387503444300272 Chr1 123 124 -0.822481891189735 Chr1 124 125 -0.651786473358111 Chr1 125 126 0.404937218146016
Note: This file should be sorted by coordinate (sort -k 1,1 -k 2,2n).
Tip: Due to the PhyloP file may be very large, it is recommended to split it and run FunTFBS for each chromosome.
This file can be generated by multiple alignments or downloaded from UCSC genome browser.
- Genome sequences in fasta format, for example:
>Chr1 CCCTAAACCCTAAACCCTAAACCCTAAACCTCTGAATCCTTAATCCCTAAATCCCTAAATCTTTAAATCC TACATCCATGAATCCCTAAATACCTAATTCCCTAAACCCGAAACCGGTTTCTCTGGTTGAAAATCATTGT GTATATAATGATAATTTTATCGTTTTTATGTAATTGCTTATTGTTGTGTGTAGATTTTTTAAAAATATCA TTTGAGGTCAATACAAATCCTATTTCTTGTGGTTTTCTTTCCTTCACTTAGCTATGGATGGTTTATCTTC ATTTGTTATATTGGATACAAGCTTTGCTACGATCTACATTTGGGAATGTGAGTCTCTTATTGTAACCTTA GGGTTGGTTTATCTCAAGAATCTTATTAATTGTTTGGACTGTTTATGTTTGGACATTTATTGTCATTCTT ACTCCTTTGTGGAAATGTTTGTTCTATCAA
This file can be generated by genome sequencing or downloaded from genome database.
Here is a simple example using the demo files in the package:
funTFBS -t demo/test_TFBS.bed -m demo/Ath.meme -f meme -p demo/test_PhyloP.bed -g demo/Ath_test.fa -o test
Following information will be shown on the screen:
TFBS before filtering: 100 motif format: meme TFBS after filtering: 20 test/TFBS_filtered.bed
After running it there will be two files generated in the output directory, which are in bed6+ format (9 columns).
TFBS_unfiltered.bed: Total candidate TFBS before filtering. TFBS_filtered.bed: Functional TFBS after filtering.
The 9 columns of output files:
| 1 | Chromosome | The chromosome name |
| 2 | Start | The starting position of the TFBS |
| 3 | End | The ending position of the TFBS |
| 4 | TF | The TF ID |
| 5 | Value | (kept from input file and not used) |
| 6 | Strand | The strand information of the TFBS |
| 7 | Sequence | The genomic sequence of the TFBS |
| 8 | Correlation | Pearson correlation between motif base frequencies and the absolute value of PhyloP |
| 9 | P-value | P-value in the correlation test |
For example:
Chr1 8649 8666 AT4G18450 3.51e-06 - ATGGCGGCGAGTGAACA 0.5576 2.003e-02 Chr1 8650 8671 AT1G72360 8.17e-07 + GTTCACTCGCCGCCATTGCTC 0.5802 5.829e-03 Chr1 8650 8671 AT3G11020 4.61e-06 - GAGCAATGGCGGCGAGTGAAC 0.5283 1.382e-02 Chr1 8650 8671 AT4G17490 7.99e-07 + GTTCACTCGCCGCCATTGCTC 0.5882 5.042e-03
1) Source of TF binding motifs
PlantTFDB - Plant Transcription Factor Database
JASPAR - The high-quality transcription factor binding profile database
UniPROBE - Universal PBM Resource for Oligonucleotide Binding Evaluation
CIS-BP - The online library of transcription factors and their DNA binding motifs
TRANSFAC - The database of eukaryotic transcription factors
New PLACE - A Database of Plant Cis-acting Regulatory DNA Elements
PlantPAN - The Plant Promoter Analysis Navigator
PlantCistromeDB - Base-pair Resolution Atlases of the Plant Cistrome and Epicistrome
2) Source of sequence conservation scores
PlantRegMap
UCSC Genome Browser
3) Dataset used to evaluate different methods in Arabidopsis thaliana
Binding sites and regulations for 21 TFs in different levels: Download
High-confidence transcriptional regulatory map: ATRM
Gene ontology slim: TAIR
Gene expression correlation: ATTED-II
4) Functional TFBS and regulations generated by FunTFBS for 63 species
Explore predicted TFBS in genome browser
Download predicted TFBS
Retrive regulations on-line
Download regulations
How To Cite
- Tian F, Yang DC, Meng YQ, Jin JP and Gao G. (2019). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Research, gkz1020.



